How to organize your grants using machine learning
Categories:
Large grant giving organizations like the Wellcome Trust or Cancer Research UK receive thousands of applications for funding grants each year. Analysis of this data can be complicated by the fact that adding useful metadata to funding grant applications is often time consuming to do manually.
By metadata, we mean information such as what diseases was the applicant interested to study, or what part of the body is targeted by a new drug proposed in a grant? This kind of information, which would be very useful to guide later analysis, will almost certainly be captured in the grant title or summary, but it may not be captured implicitly in the metadata in a way that can be simply queried.
Link to demo https://huggingface.co/spaces/mantisnlp/SearchMesh
It can, in fact, be difficult for a funding organization to know in advance all of the ways that they would want to break down their grants. Even if they do know, it might require specialist expertise to add that information to every grant. This is not a problem that is relevant only to large grant giving organizations. Smaller ones also have the need to extract similar insights and they often have to do it ad hoc each time they want to analyze their grants; they may manually define the categories of interest, possibly in slightly different ways.
This is the problem we worked with the Wellcome Trust to resolve: how can we use machine learning to better organize their grants. The first step was to agree on a classification system, i.e. a set of relevant labels that we could apply to the grants. There are a couple of research classification systems such as fields of research (FOR) which includes major fields and sub-fields of research. The Wellcome Trust preferred a biomedically focused system that contained a large enough set of labels to cover the analytical needs of the Trust for years to come. We settled on Medical Subject Headings (MeSH), a classification system developed by the National Library of Medicine (NLM). MeSH contains twenty nine thousand tags and it is being widely used so it would allow Wellcome to make comparisons with other funders that are also using it.
In order to train a machine learning algorithm to classify grants with MeSH, we needed a dataset of biomedical related documents that had already been labeled with MeSH. Fortunately, most biomedical publications are annotated with MeSH tags by expert annotators from the National Library of Medicine (NLM) when they are indexed in their database (MEDLINE) which can be accessed through PubMed. Within this corpus of biomedical publications, there are more than fourteen million publications tagged with one or more of twenty nine thousand MeSH tags.
Training a machine learning algorithm to learn to apply MeSH tags to Wellcome grants that it has not seen before was technically challenging both due to the number of documents in our training dataset, and the number of tags that we need a machine learning algorithm to learn to apply. This problem is called extreme multilabel classification. Extreme because there are tens of thousands of tags (an order of magnitude more than most machine learning algorithms), and multilabel because each grant can have more than one MeSH tag applied to it.
In trying to solve this problem we tried a range of different techniques from traditional natural language processing (NLP) approaches, such as tfidf svm, to the latest state-of-the-art transformer models. We ended up with a tradeoff between a fast 🏎 and less performant linear model and a slower 🐌 but much more accurate neural network 🤖.
Our linear model takes less than 4 hours to be trained using 16 CPUs while achieving 56% accuracy, whereas our transformers based model takes 4 days to train using 8 GPUs achieving 63% accuracy. It is worth noting the state of the art (SOTA) is around 70% but the associated models are not openly available. Predicting tags on Wellcome grants is much quicker since there are much fewer grants than biomedical publications.
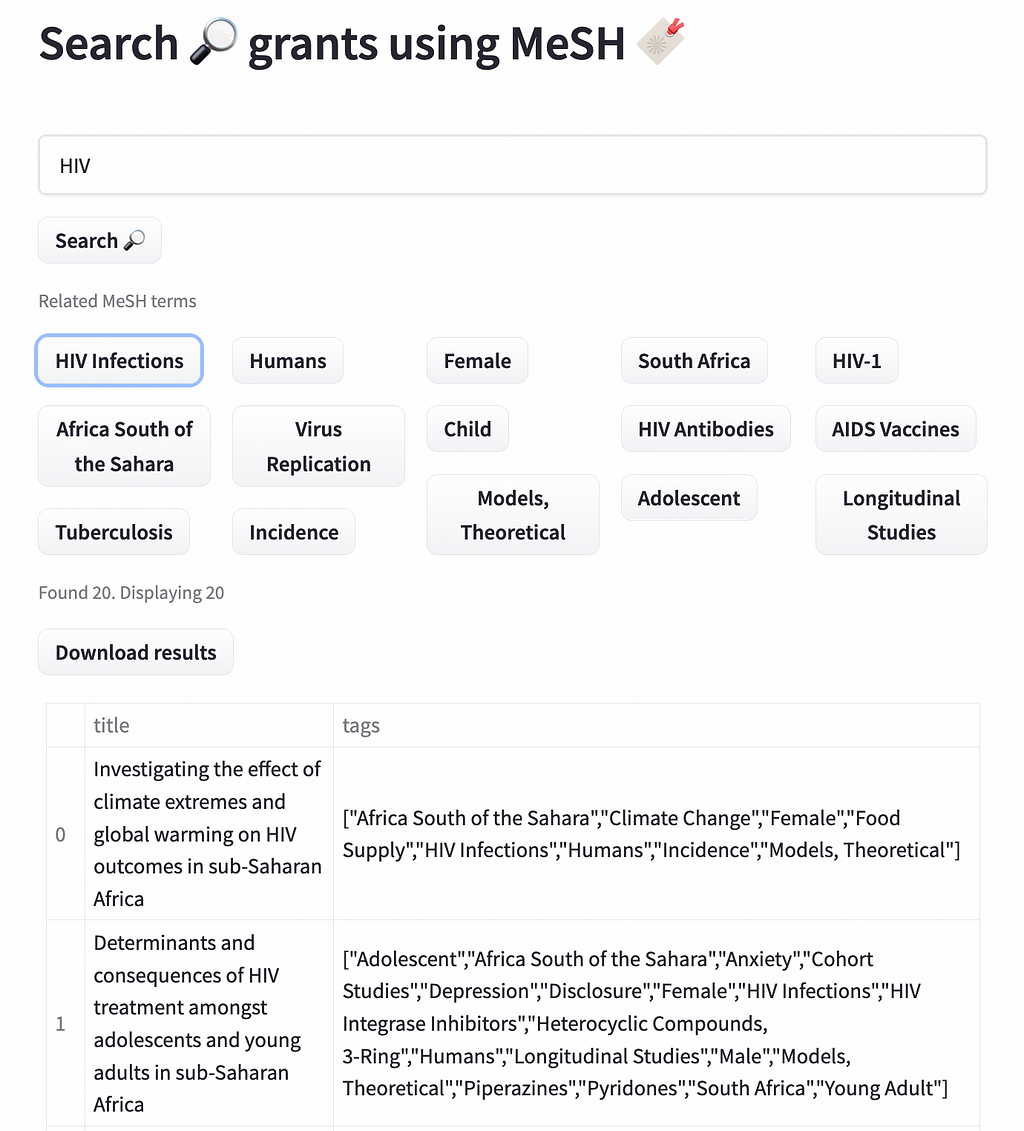
Adding MeSH tags to Wellcome grants opens the door to answering many new analytical questions that could not have been easily answered before. These questions can also now be asked in a more standardized way, no time is lost in defining concepts such as malaria or mental health each time an analyst needs to retrieve related grants. They can also be as prescriptive as they want, for example retrieving only grants for ‘malaria vivax’ (the most common species of malaria) or a specific anxiety disorder such as ‘separation anxiety’.
We created a demo of how those tags can be used to search Wellcome grants. The data is based on the openly available data through 360giving. You can access the demo here https://huggingface.co/spaces/mantisnlp/SearchMesh
We have also created a space where you can use the model to predict MeSH tags in your own grants. You can access that here https://huggingface.co/spaces/mantisnlp/MeshTagger
Hopefully, this paints the picture of how machine learning can be used to automate an otherwise expensive and manual process and open the possibility for extracting additional value from your existing grants data. If you have any questions or you are interested to apply this in your organization feel free to reach out to us at hi@mantisnlp.com.
How to organize your grants using machine learning 🏷 was originally published in MantisNLP on Medium, where people are continuing the conversation by highlighting and responding to this story.